Engineered Enzymes for Membrane Editing
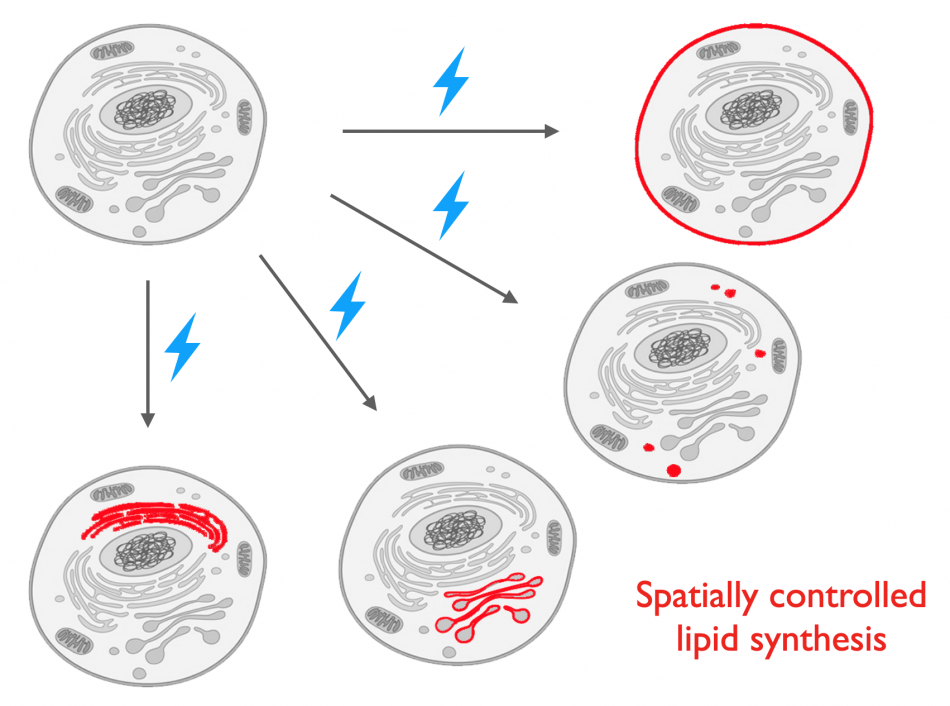
Individual lipids can signal from numerous places in the cell, and the same lipid can deliver different messages depending on where it is positioned. Yet, for many lipids, it is not well understood how the spatial context where they are generated can dictate specific signaling outcomes in the cell. Thus, to complement our tools for visualizing the spatial locations where endogenous lipid signaling occurs, we are developing distinct approaches for manipulating lipid signaling with spatiotemporal precision, an approach we have termed membrane editing.
We have begun by focusing our efforts on phosphatidic acid (PA), a potent and multifunctional phospholipid that controls many pathways in the cell, including actin cytoskeletal dynamics, membrane trafficking, and proliferation, and whose production is dysregulated in many cancers. We have designed enzyme-based tools that resides within cells in an inactive form but, with a rapid and non-toxic stimulus, activates phosphatidic acid production on specific organelle membranes.
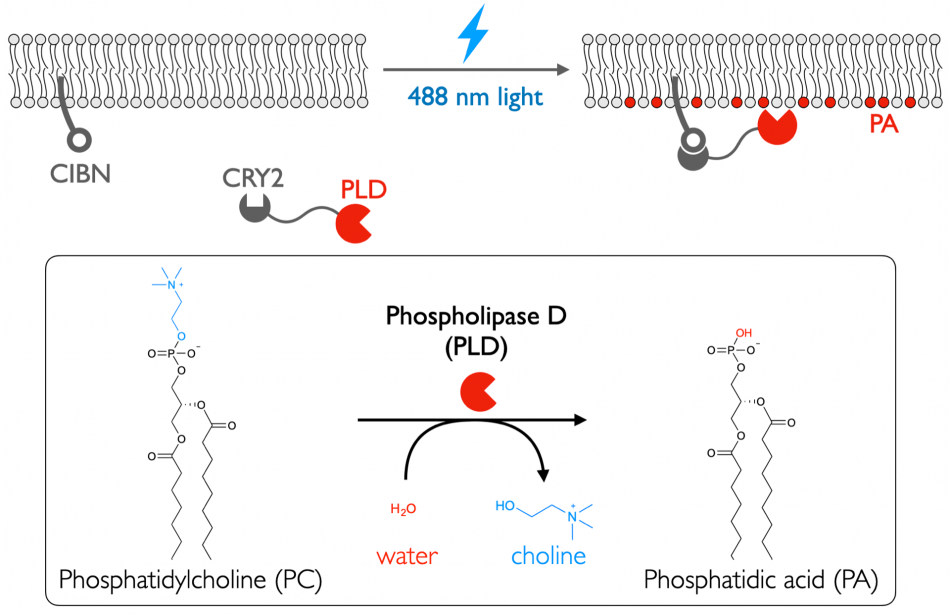
Our tools, termed optoPLDs for optogenetic phospholipase Ds (PLDs), involve the fusion of an engineered, microbial PLD to one half of the light-mediated CRY2–CIBN heterodimerization system, while the other half is constitutively targeted to a specific organelle membrane. Following brief illumination with blue light, this optoPLD is recruited to the organelle membrane of interest, leading to spatially restricted generation of PA. We have combined our IMPACT tools for visualizing PLD activity with directed evolution to identify variants of PLD with altered catalytic efficiencies to modulate the amplitude of PA signaling induced by optoPLD. We are continuing these directed evolution efforts to create unnatural PLD enzymes with unique properties to generate an expanded set of synthetic signaling circuits, including super-active “superPLDs”. Recently, we have designed next-generation, ultralow background optogenetic membrane editors termed LOVPLDs that enable turn-on of PLD catalytic activity rather than change in localization. We are applying our membrane editors to elucidate mechanisms controlling cellular PA homeostasis and roles for PA signaling in growth and proliferative pathways implicated in cancer.
Key Publications
Chen PB*, Li XL*, Baskin JM. “Synthetic Lipid Biology.” Chem Rev (2025) 125, 2502-2560. *Equal contribution.
Tei R, Baskin JM. “Dynamic network regulating phosphatidic acid homeostasis revealed using membrane editing coupled to proximity labeling.” bioRxiv (2024). doi: https://doi.org/10.1101/2024.09.14.612979.
Li, X-L, Tei R, Uematsu M, Baskin JM. “Ultralow background membrane editors for spatiotemporal control of lipid metabolism and signaling.” ACS Cent Sci (2024) 10, 3, 543–554. (Highlighted in Nat Chem Biol and ACS Cent Sci)
Tei R, Bagde SR, Fromme JC, Baskin JM. “Activity-based directed evolution of a membrane editor in mammalian cells.” Nat Chem (2023) 15, 1030–1039.
Tei R and Baskin JM. “Spatiotemporal Control of Phosphatidic Acid Signaling with Optogenetic, Engineered Phospholipase Ds.” J Cell Biol (2020) 219, 3, e201907013.

