Chemical Approaches for Imaging Lipid Signaling
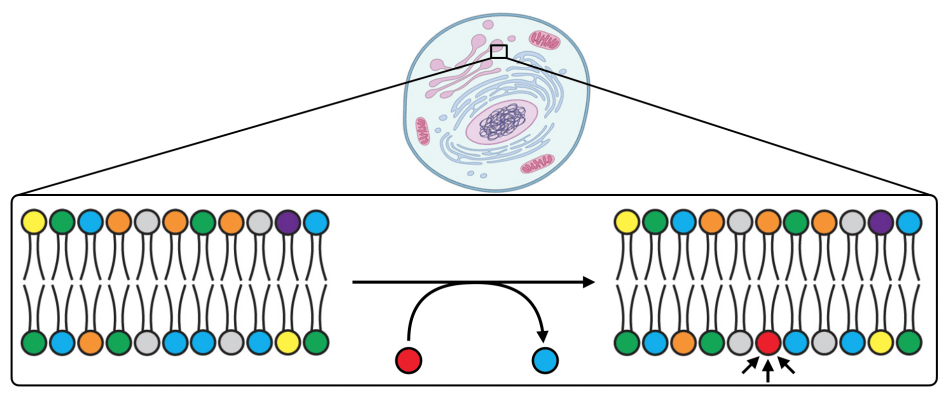
Every organelle within every cell in our body has a unique lipid composition in its membranes. This lipid composition can change when cells respond to environmental stimuli, as certain lipids are produced to act as signaling agents (red in the figure above). Because these lipid signaling agents can elicit numerous downstream physiological outcomes, it is a great mystery how specificity can be achieved. The location where a particular lipid is generated is a critical and often overlooked property, beyond its molecular identity, for dictating specific signaling events. Yet, for many lipids, tools do not yet exist for accurately visualizing their production and dynamics within live cells. We are developing molecular technologies for imaging the production of important signaling lipids.
In particular, we have focused our efforts on phosphatidic acid. This potent and multifunctional phospholipid controls many pathways in the cell, including actin cytoskeletal dynamics, membrane trafficking, and proliferation, and its production is dysregulated in many cancers. A major route that cells use to synthesize phosphatidic acid is hydrolysis of abundant membrane phospholipids by phospholipase D (PLD) enzymes. We have developed a suite of small-molecule tools for visualizing sites of phosphatidic acid production by PLD enzymes within live cells.
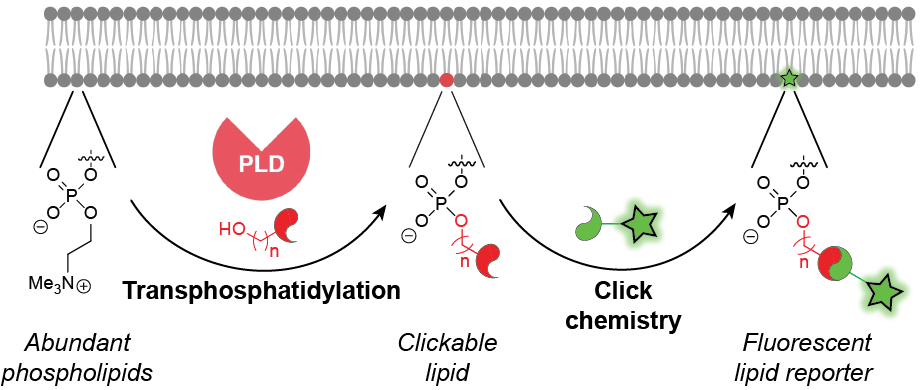
Our toolset, termed IMPACT, for imaging phospholipase D activity with clickable alcohols via transphosphatidylation, relies upon the catalytic promiscuity of PLD enzymes. We have discovered that, in addition to using water for phospholipid hydrolysis, PLD enzymes accept a wide variety of synthetic, bioorthogonally tagged primary alcohols in a transphosphatidylation reaction. Subsequent tagging of the resultant clickable lipids via a bioorthogonal click chemistry reaction yields fluorescent lipids that act as reporters of PLD activity. These fluorescent lipids can be monitored by time-lapse, live-cell fluorescence microscopy to reveal subcellular locations of PLD-mediated phosphatidic acid signaling in response to different physiological stimuli of these pathways. Additionally, they can enable visualization of intracellular lipid transport pathways. IMPACT also enables quantification of PLD signaling activity at the single-cell level by flow cytometry and bulk biochemical level by HPLC and LC–MS. We are currently applying IMPACT to elucidate new functions for PLD and phosphatidic acid signaling in the regulation of numerous physiological and pathological events.
Key Recent Publications
Liang D, Luan L, Baskin JM. “Imaging interorganelle phospholipid transport by extended synaptotagmins using bioorthogonally tagged lipids.” bioRxiv (2023). doi: https://doi.org/10.1101/2023.02.01.526230.
White BM, Kumar P, Conwell AN*, Wu K*, Baskin JM. “Lipid expansion microscopy.” J Am Chem Soc (2022) 144, 40, 18212–18217. *Equal contribution.
Chiu D-C, Baskin JM. “Organelle-selective membrane labeling through PLD-mediated transphosphatidylation.” JACS Au (2022) 2, 12, 2703–2713.
Yu W, Lin Z, Woo CM, and Baskin JM. “A chemoproteomics approach to profile phospholipase D-derived phosphatidyl alcohol interactions.” ACS Chem Biol (2022) 17, 12, 3276–3283. Part of Young Investigator special issue.
Bumpus TW, Huang S, Tei R, and Baskin JM. “Click chemistry–enabled CRISPR screening reveals GSK3 as a regulator of PLD signaling.” Proc Natl Acad Sci USA (2021) 118, 48, e2025265118.
Liang D, Cheloha R, Watanabe T, Gardella TJ, and Baskin JM. “Activity-based, bioorthogonal imaging of phospholipase D reveals spatiotemporal dynamics of GPCR-Gq signaling.” Cell Chem Biol (2021) 28, 1–7.
Liang D, Wu K*, Tei R*, Bumpus TW, Ye J, and Baskin JM. “A real-time, click chemistry imaging approach reveals stimulus-specific subcellular locations of phospholipase D activity.” Proc Natl Acad Sci USA (2019), 116, 31, 15453–62.

